ImmUniverse IDs#
To manage the complex data landscape of the ImmUniverse project, a consortium wide identifier re-pseudonymization scheme is developed, which uniquely tags every study participant, each collected biospecimen, and its assay by ImmUniverse_SUBJECT_ID, and ImmUniverse_SAMPLE_IDrespectively.
Overview of the ImmUniverse ID assignment#
- Each study participant in ImmUniverse is tagged with a unique
ImmUniverse_SUBJECT_ID - Each collected biospecimen is tagged with a unique
ImmUniverse_SAMPLE_IDwhich contains theImmUniverse_SUBJECT_IDto connect it to the participant
The structure of ImmUniverse IDs is illustrated with an example in Figure 1.
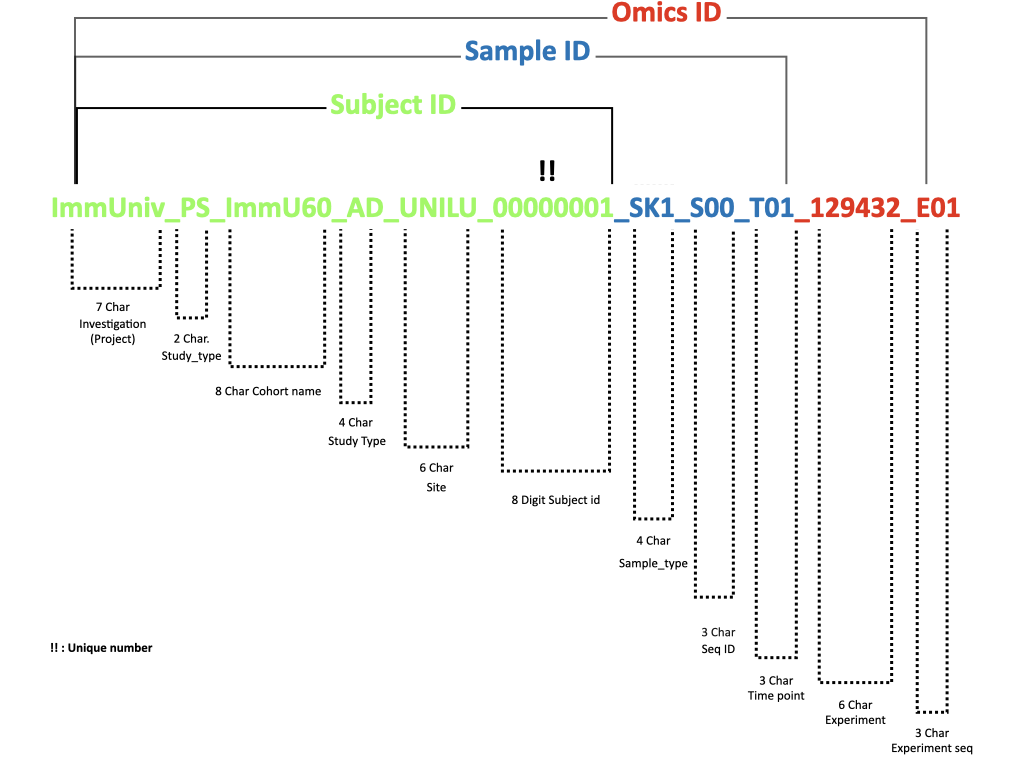 Figure 1: Structure of ImmUniverse IDs
Figure 1: Structure of ImmUniverse IDs
In the ImmUniverse Data Platform, it is by design mandatory to tag each study participant with ImmUniverse_SUBJECT_ID; each biospecimen with ImmUniverse_SAMPLE_ID.
ImmUniverse data uploaders are responsible to generate ImmUniverse IDs at their location before uploading the data to UNILU, and they will store a map between the ImmUniverse IDs and their internal IDs. The uploaded data should only contain the ImmUniverse IDs, not the internal IDs.
ImmUniverse_SUBJECT_ID - unique subject identifier#
Every study participant in the ImmUniverse consortium is assigned with a unique ImmUniverse_SUBJECT_ID. It should be generated at the data uploaders’ location by re-pseudonymization following a common structure.
ImmUniverse_SUBJECT_ID consists of five parts - PROJECT_NAME , COHORT_NAME, COHORT_TYPE, SITE_OF_RECRUITMENT, and SUBJECT_NO delimited by _ (underscore) (example: IMMUNIV_ImmU_AD_UNILU_00000001).
PROJECT_NAME- the name of the project (alwaysIMMUNIV)STUDY_TYPE- Type of the study in ImmUniverse- a code from a controlled vocabulary list (example:PS).COHORT_NAME- the short name or acronym of the cohort (example:ImmU)SUBJECT_TYPE- the short name or subject of the cohort (example:AD)SITE_OF_RECRUITMENT- the institute code of the site of recruitment or cohort custodian (example:UNILU).SUBJECT_NO- an eight-digit re-pseudonymized subject identifier (example:00000001).SUBJECT_NOis not required to be sequential.
For example, the subject identifier IMMUNIV_PS_ImmU_AD_UNILU_00000001 refers to the study participant with number as ‘0000001’ in the ImmU cohort who was recruited at UNILU for the cohort AD. The cold and description of SUBJECT_TYPE and SITE_OF_RECRUITMENT could be found in Table 1.
Table 1: SUBJECT_TYPE and SITE_OF_RECRUITMENT codes and description
Note: UNILU does not provide any sample and this is only an example.
ImmUniverse_SAMPLE_ID - unique sample identifier#
Each collected biospecimen or sample in ImmUniverse requires a re-pseudonymized ImmUniv_SAMPLE_ID which extends the ImmUnive_SUBJECT_ID with SAMPLE_TYPE, and STUDY_TYPE SMPLE_SEQ and TIME_POINT delimited by _ (underscore).
SAMPLE_TYPE- Type of the specimen - a code from a controlled vocabulary list (Table 2).SMPLE_SEQ- A sequence number to clarify repeated/replicated/duplicated/aliquoted sampling.TIME_POINT- Cohort or study-specific identifier for visit(s) in longitudinal study. Example:T01,T02for sampling repetitions;TNNif there is no sampling repetition.
For example, the sample identifier IMMUNIV_PS_ImmU_AD_UNILU_00000001_SK1_S01_T04 refers to the aliquot as 01 of the skin sample from the Prospective study collected at the time-point 04 from sample MMUNIV_PS_ImmU_AD_UNILU_00000001_SK1. Here, ‘PS’ is used to represent for ‘Prospective study’ and RS for ‘Retrospective study’. The cold and description of TIME_POINT and SAMPLE_TYPE could be found from in Table 2.
Table 2: SAMPLE_TYPE and TIME_POINT codes and description
ImmUniverse_OMICS_ID - unique omics data identifier#
All omics data generated from the ImmUniverse requires a Immuniverse_omics_ID, which has three parts: IMMNIV_SAMPLE_ID, OMICS_TYPE and OMICS_SEQ delimited by _ (underscore)
OMICS_TYPE- Type of the assay; UNILU strongly recommends using using NCIT or BioAssay Ontology (BAO) termsOMICS_SEQ- Sequence number or alpha numeric code for the repeat assays (example: E00,E01 )
For example, the data or assay identifier IMMUNIV_PS_ImmU_AD_UNILU_00000001_SK1_S01_T04_129432_E01 refers to the RNA-seq assay which was done on the sample IMMUNIV_PS_ImmU_AD_UNILU_00000001_SK1_S01_T04 for an assay repeat sequence E01. The output data files and omics metadata of this assay will be named/taggged with IMMUNIV_PS_ImmU_AD_UNILU_00000001_SK1_S01_T04_129432_E01.
If the assay is not repeated ASSAY_SEQ should be assigned to E00.
You can download the summary of the code and its description as an Excel file.
Please contact us if you need help with renaming or formatting your high-dimensional data files.
