Prepare data for uploading#
This document provides instructions on how to prepare the clinical and omics data.
Data should be organised and formatted to a common standard before uploading onto the ImmUniverse data platform and computing environment.
Steps and requirements for accessing the data upload link#
Prior to uploading any clinical or molecular data files, it is mandatory to comply with the ImmUniverse’s legal requirements and provide data protection information. In addition to the requirements outlined herein, you may be subject to local data protection laws, clinical trial regulations or local ethics requirements. Therefore, prior to uploading any data, it is advised that you speak with your data protection officer (DPO) to understand the full scope of your requirements. For public domain data, go to Section 3.
1. Data Sharing Agreement#
Make sure that the upload and sharing of the dataset(s) in question is covered by the ImmUniverse consortium Data Sharing Agreement (DSA). Each partner in ImmUniverse has been asked to complete the DSA, so please check with your institutions lead member to confirm.
If the dataset is not covered by the DSA, a separate agreement covering articles 26 & 28 needs to be signed between at least the UNILU and the uploading partner.
Before uploading any data, please speak with your DPO to ensure they are kept aware of, and agree with, the procedure for uploading your data as outlined in the Data Upload Instructions section.
2. UNILU Data Protection Information Sheet#
DISH will allow you to document use restrictions on the cohort data you are uploading. Make sure that you document on this sheet the list of parties that should not receive access to the data or for whom access restrictions need to be implemented. Please download the DISH template with the following link:
3. Re-pseudonymization data#
In the ImmUniverse Data Platform, it is by design mandatory to tag each study participant with ImmUniverse_SUBJECT_ID; each biospecimen with ImmUniverse_SAMPLE_ID; and each experimental procedure or output with ImmUniverse_EXPERIMENT_ID/ImmUniverse_OMICS_ID.
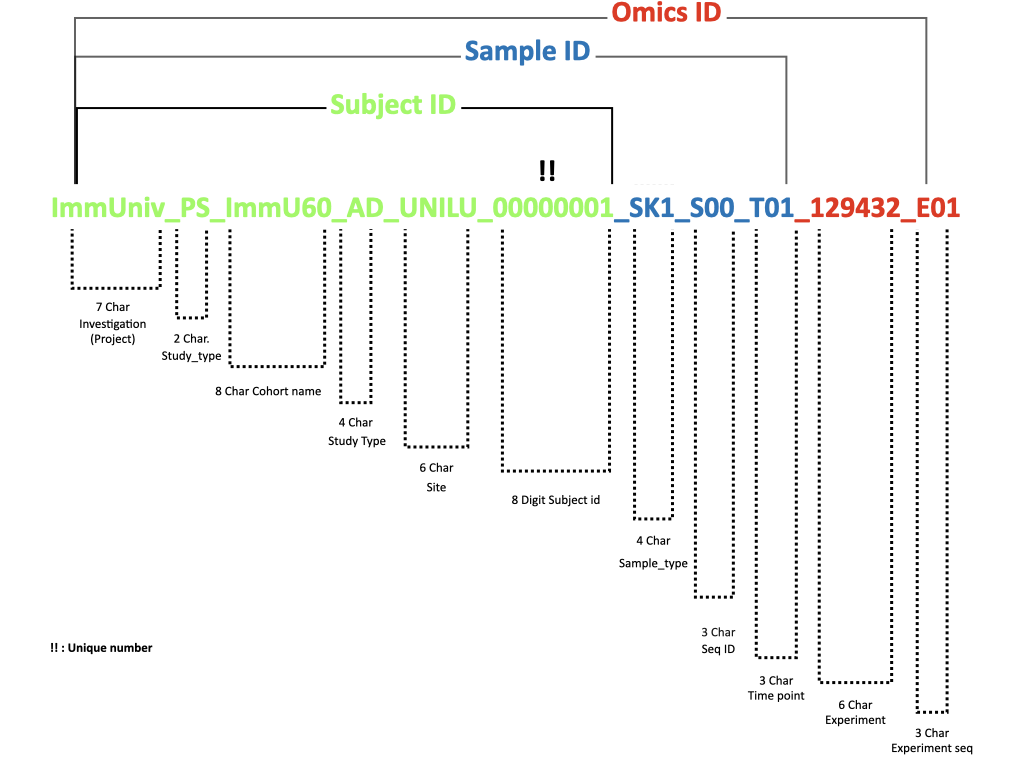
Read more about ImmUniverse IDs.
To rename the data with ImmUniverse IDs, please download this template for renaming the data or access the IDs generation REDCap to get more details.
Do you need more help to format your data? Please contact us.
4. Directory structure#
Please follow the steps below to organise your local directory in order to be compliant:
- The name of the top level folder should contain the
cohort nameand the upload date stamp inYYYYMMDDformat, separated by_. Eg -20221103_UNILU_ImmU60 - Directory structure inside the top level folder should be consistent with the data-types -
clinical,genomics,transcriptomics,Methylomics,single cell,microbiome,RNA-seq,Auto-antibody,miRMA,Immunophenptypedata,Proteomics,Metabolomics,Lipdomics. - Sub_directory should be “raw-data” and “processed-data”
- If your data contains any other types of data not mentioned, add them in a directory named
others - Inside the
othersdirectory create a separate directory for each of such data types - Add a
READMEfile in the plain-text or markdown format with a description of the uploaded data, and any other relevant information - Multiple small files should be put into a file archive, such as zip or tar for faster upload.
- On the clinical data, please use the template.
An example of the compliant directory structure:
+ 20221103_UNILU_ImmU60
|- - + clinical
| |- - clinical_data.tsv
|
|- - + genomics
| |- - + raw-data
| | |- - genomics_metadata.tsv
| | |- - genomics_rawdata.zip
| |- - + processed-data
| | |- - genomics_metadata.tsv
| | |- - genomics_rawdata.zip
|
|- - + others
| |- - + imaging_data
| | |- - imaging_data.zip
|
|- - README
Do you need more help to format your data? Please contact us.
5. Naming conventions for experimental output data#
To link experimental data to clinical as well as experimental metadata with a unique naming of the output files resulting from the experiment is required. Accordingly, renaming the files employing the ImmUniverse_OMICS_ID is mandatory. In the case of experiment-specific ambiguities, we suggest using the manufacturer’s file naming conventions.
Example: paired-end sequencing experiments result in one output for each of the reads R1 and R2. We assume raw sequencing files are named in the following way:
ImmUniv_ImmU60_AD_UNILU_00000001_PS_SK1_S00_T01_129432_E01_R1.fastq.gz
ImmUniv_ImmU60_AD_UNILU_00000001_PS_SK1_S00_T01_129432_E01_R2.fastq.gz
The UNILU will rename the data automatically according the ImmUniverse ID mapping table you filled in Section 3.
Please contact us if you need help with renaming your experimental output files.
6. Data formatting#
All files must be machine readable and use a common data format:
Clinical data should be provided as tab-separated values (TSV) preferably. In case you cannot make your data available as TSV, please get in touch with the UNILU Team. Numeric data not consisting of whole numbers must be represented according to the XML float lexical representation sepcifications. In short, the decimal separator must be a dot
., and large numbers are represented in “E notation”.Clinical data must be structured in accordance with the data dictionary (see ImmUniverse metadata upload guide for details), i.e. column names must match the variable names in the data dictionary, and values are of the types, ranges, units as described in the data dictionary.
Omics data should be provided in the most appropriate file format, e.g. FASTQ for RNA-seq. Omics data must be accompanied by metadata. See ImmUniverse metadata upload guide for details.
More help#
Do you need more help to format your data? Please contact us.
